MOLPRO
MOLPRO - Ab-initio programs for molecular electronic structure calculations
Introduction
Molpro is a complete system of ab initio programs for molecular electronic structure calculations, designed and maintained by the following authors:
Hans-Joachim Werner | Peter Knowles |
As distinct from other commonly used quantum chemistry packages, the emphasis is on highly accurate computations, with extensive treatment of the electron correlation problem through the multi-configuration-reference CI, coupled cluster and associated methods.For further details and documentation, please consult the MOLPRO Home page.
Licensing and versions
The Molpro software and documentation is copyright material owned by University College Cardiff Consultants Limited and is distributed under license to academic users of LRZ.
Usage
Molpro Input
The MOLPRO User's manual contains a detailed description of MOLPRO input.
Running Molpro interactively at LRZ
Before running molpro jobs interactively please load the appropriate module environment via:
module load molpro
To see where the molpro executables reside (the bin directory) and what environment variables it defines, type
module show molpro
The execution of Molpro can be done as follows:First, salloc is invoked to reserve the needed resources.
molpro [-n N] inputfilename (N: number of processors)
Running parallel Molpro using job scripts at LRZ
To use Molpro in batch mode, please find below example job scripts for all the available platforms:
Linux-Cluster and SuperMUC-NG (SLURM) |
|---|
#!/bin/bash #SBATCH -o ./%x.%j.%N.out #SBATCH -D ./ #SBATCH -J <job_name> #SBATCH --clusters=cm2_tiny |
Then submit the job script using sbatch (SLURM) or llsubmit (LoadLeveler) commands.
e.g., assume the job script name is name-job.pbs:
% sbatch name-job.pbs
Memory Requirements
The serial MOLPRO executable knows one kind of memory.
Its amount is specified by the -m command line option to the molpro command, or the MEMORY card in the Molpro input file.
molpro -m 8M
means 8 MWords = 8*1024*1024 Words = 8388608 8-byte words = 64 Mbyte
When running parallel MOLPRO, the amount of the GA memory has to be added to the product of number of tasks and task memory to obtain the total memory need of the run. Running 32 tasks on one node with defaults of
molpro -m 8M -G 8M
yields 32 * 8 MW + 8 MW = 2112 MByte total memory consumption on this node.
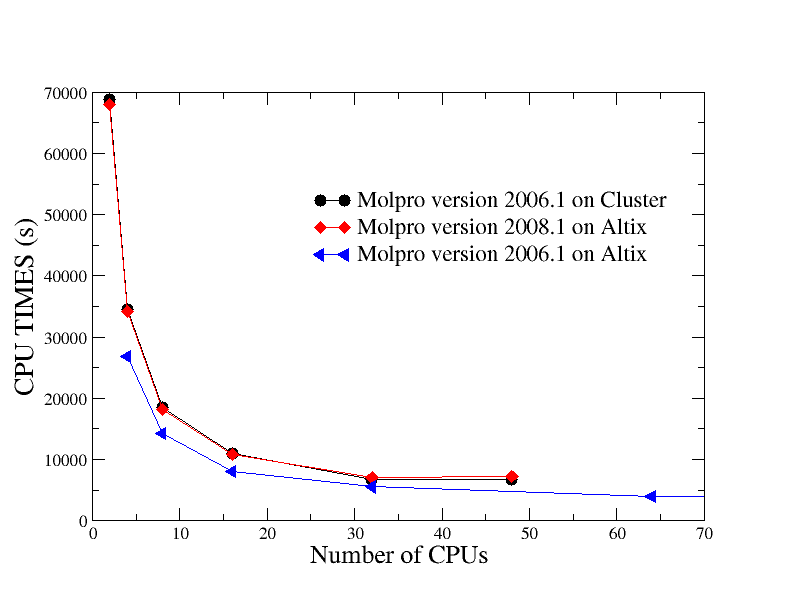
Example of strong scaling on Cluster and Altix
The used input is CrCN with memory of 130m. The performance results up to 128 cores on Altix and up 48 cores on Cluster dataset are plotted in Figure below.
Support at LRZ
If you have any questions or problems with the installed Molpro versions on the different LRZ platforms, please don't hesitate to contact:
Dr. M. Allalen (allalen_at_lrz.de)