Julia for HPC
Julia is, similar to Python and R, a very user oriented, flexible tool for computing and software development. But it appears very easy to be installed and adapted by users. Out of this reason, we do not provide a central julia installation. Still, a lot of questions appear about the usage of Julia on SuperMUC-NG. These are tried to get answered here. If something is missing, please open a service request on our Service Desk!
Generally, the Julia Docu is a good point to obtain help!
Getting started
> module av julia
----------- /lrz/sys/share/modules/files_sles15/tools -------------
julia/1.5.4 julia/1.6.5_lts julia/1.7.0 julia/1.7.1 julia/1.7.2
> module show julia/1.7.2
-------------------------------------------------------------------
/lrz/sys/share/modules/files_sles15/tools/julia/1.7.2:
module-whatis {Julia: Programming Framework}
[...]
-------------------------------------------------------------------
> module load julia/1.7.2
We provide some basic modules centrally such as ClusterManagers, MPI, Plots and BenchmarkingTools. If more modules are centrally needed, please open a Service Request. Generally, however, users can load/install own packages (even project specific if needed). So, we try to minimize the number of centrally installed packages in order to avoid version requirement clashes. That's specifically true as Julia allows for a rather complex and dynamic package handling for each project, up to even including complete conda environments.
Installing Julia
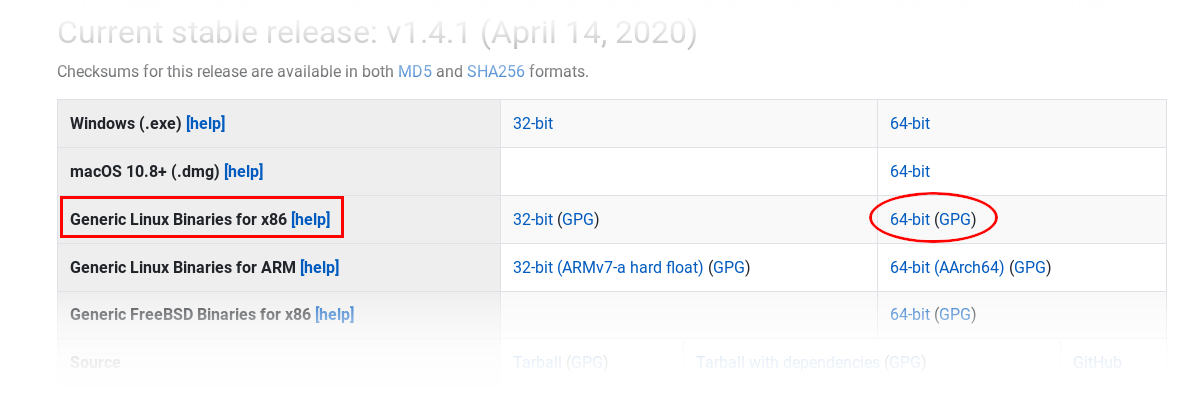
The easiest way to get your own Julia into operation is via the download of the binary packages from the Julia Download Site.
Use a SCP client of your choice to upload the downloaded file to your SuperMUC-NG account (scp, pscp, WinSCP, Filezilla, ...).
> tar xf julia-1.4.1-linux-x86_64.tar.gz
> echo 'export PATH=$PATH:$HOME/julia-1.4.1/bin' >> ~/.profile && source ~/.profile # or add bin otherwise to your search path, e.g. via alias!
> julia
_
_ _ _(_)_ | Documentation: https://docs.julialang.org
(_) | (_) (_) |
_ _ _| |_ __ _ | Type "?" for help, "]?" for Pkg help.
| | | | | | |/ _` | |
| | |_| | | | (_| | | Version 1.4.1 (2020-04-14)
_/ |\__'_|_|_|\__'_| | Official https://julialang.org/ release
|__/ |
julia>
Please modify accordingly if you have a different install path! Newer versions should work the same way.
That's it. You can go on!
Installing/Updating Julia Packages (SuperMUC-NG-specific)
As with Python and R, also Julia offers a dynamic package development, deployment and managing concept, which usually requires a direct access to the Internet. SuperMUC-NG does not offer this feature. So, you must use a workaround. The same local Python HTTP proxy - reverse SSH tunnel method, as described for Conda, also works here.
This procedure is only necessary when you want to add packages! Usual execution of Julia code does not require any further package download, or SSH tunnel, or Python HTTP Proxy!
Preparation (local PC/Laptop)
Setup a local HTTP Proxy using Python:
local> pip3 install --upgrade --user proxy.py local> ~/.local/bin/proxy --port 3128 # the port number is arbitrary, but you must remember it!
Next, open a SSH connection to SuperMUC-NG, with a reverse tunnel:
local> ssh -l <your-lrz-account-id> -R 3128:localhost:3128 skx.supermuc.lrz.de # left port number must agree with that of the HTTP proxy above! sng>
Preparation (SuperMUC-NG)
In a next step, create the file ~/.julia/config/startup.jl and insert the two lines
ENV["HTTP_PROXY"] = "localhost:3128" ENV["HTTPS_PROXY"] = "localhost:3128"
Take care to use the same port number as on the right of the SSH reverse tunnel above!
Package Installation
Next, you should simply start julia on the SuperMUC-NG login node and press ']' key to switch to the package manager. It should be as simple as issueing add Plots, for instance.
Unresolved Issue: The initial update will fail. So, also the installation of any package! The error is probably something like:
ERROR: failed to clone from https://github.com/JuliaRegistries/General.git, error: GitError(Code:ERROR, Class:Net, unrecognized URL prefix)
This is related to the download of the Julia registry (which is quite large, btw.). This must be done manually, too.
> mkdir -p .julia/registries/ && cd .julia/registries/ ~/.julia/registries> export HTTPS_PROXY=localhost:3128 && export HTTP_PROXY=localhost:3128 # see the HTTP proxy ports above ~/.julia/registries> git clone https://github.com/JuliaRegistries/General.git ~/.julia/registries> cd General && rm -rf .git .githup && cd > julia julia> # ] (@v1.4) pkg> update ... (@v1.4) pkg> add Plots
Caution: Installation of a package may still fail at first attempt. Just retry! Also look on the output of the HTTP Proxy for error messages! It might be necessary to set export TMPDIR=$SCRATCH as the default temp folder might be to small.
Recommendation: The Julia package manager allows you to precompile your packages (also your own). Think also about whether, it is a good idea to pin the package versions.
Recommendations for Plotting
If plotting is seriously necessary in an interactive fashion on the login nodes, we recommend the use of VNC (and not X-forwarding via SSH). An overview of how to plot in Julia can be found here, for instance.
Julia comes with some plotting backends. Not all of them might work out of the box. GR, for instance, resulted in errors – see here, if you meet this problem! The solution in short – Try this:
ENV["GRDIR"] = ""
using Pkg
Pkg.build("GR")
Switching to another backend might help, too.
Parallel / Slurm
Julia exhibits several parallel paradigms – for shared memory (single node, many/multi core) and distributed (MPI). A non-comprehensive overview can you get here. The official documentation also has a chapter on parallel computing, which but does not say much about the integration into the LRZ cluster systems. This will be attempted here.
There are several parallelization paradigms available in Julia – some more explicit, others more implicit (similar as to differences in explicit thread or MPI programming, and implicit parallel library usage as in co-array Fortran). Which one meets your needs, you have to decide.
Shared Memory - OpenMP
The shared memory parallelism should work out of the box. This corresponds to to working on a single node.
Also, some packages react on OMP_NUM_THREADS, as is the case for e.g. the LinearAlgebra package.
> export OMP_NUM_THREADS=10 > julia linalg_test.jl 1.554187 seconds (2.88 M allocations: 327.748 MiB, 2.48% gc time) 0.805609 seconds (2 allocations: 190.735 MiB, 7.28% gc time) 0.813139 seconds (2 allocations: 190.735 MiB, 8.40% gc time) 0.746598 seconds (2 allocations: 190.735 MiB)
It is advisable to check the scaling behavior as a function of OpenMP threads! The linear algebra package also contains solvers for matrix equations (please check also SparseArrays)
Distributed Example - the Worker Concept with SSH Manager
julia can be started by the option -p <# of threads>, which spawn a number of threads that can be used as workers. This works only on a single node, where the name of the host does not matter. For more than one node, you must use the option --machine-file, and specify a hostname list (one name per line).
This spawns 14 workers per node according to the machine file + 1 on the local machine!.
SSH is used for this spawning. So, you have to create a passphrase-less SSH key in your ~/.ssh, and put the public key into ~/.ssh/authorized_keys.
The exercise above to add opa to each host name is in order to use the OmniPath network (on CoolMUC-2 it would be ib). Without, you get the ethernet maintenance network on SuperMUC-NG. On the Linux cluster (also on the housed systems), a similar problem exists. Please, consult the respective documentation for these systems!
Distributed Example - the Worker Concept with Slurm Manager
This example was taken from the ClusterManagers documentation page. Please have a look for more details!
In this case, no SSH keys are needed. Users are responsible for doing task/thread placement/pinning correctly via the addprocs' or addprocs_slurm's kwargs options. Doing this wrongly, can impair efficient use of the resources, and, in worst cases, cause the abort of the Slurm job.
MPI Example
This example of explicit MPI communication requires the installation of the MPI package. You can install it with the setting export JULIA_MPI_PATH=$I_MPI_ROOT, where $I_MPI_ROOT is set by the Intel MPI module, which needs to be loaded before.
More Examples and Example Use Cases
As of now there is hardly any experience with Julia at the LRZ. We would be happy to include here user feedback and examples, in order to share the experience with other – possibly new – users.
| Use case | Scripts |
|---|---|
| Julia Slurm Standalone Script with a Worker Placement Test | Check carefully the Shebang, and the |
| Simple Task Farming Scheduler (Job Farm) | As written, one can start this script with one parameter (the task database) via Instead of the external command execution, you can also directly execute functions inside the code (also python or R code, as julia allows for that e.g. via OpenMP parallel tasks are also possible. The Julia worker placement is controlled via |
Troubleshooting
If something goes wrong – specifically with the package manager – you can always start from scratch by removing ~/.julia.
Documentation and Education
[Youtube] Parallel Computing and Scientific Machine Learning
[Lauwens, Downey] Think Julia: How to Think Like a Computer Scientist
A Deep Introduction to Julia for Data Science and Scientific Computing
[SciML Tutorials] [DiffEq SciML Tutorials]